PRESS Slice Map
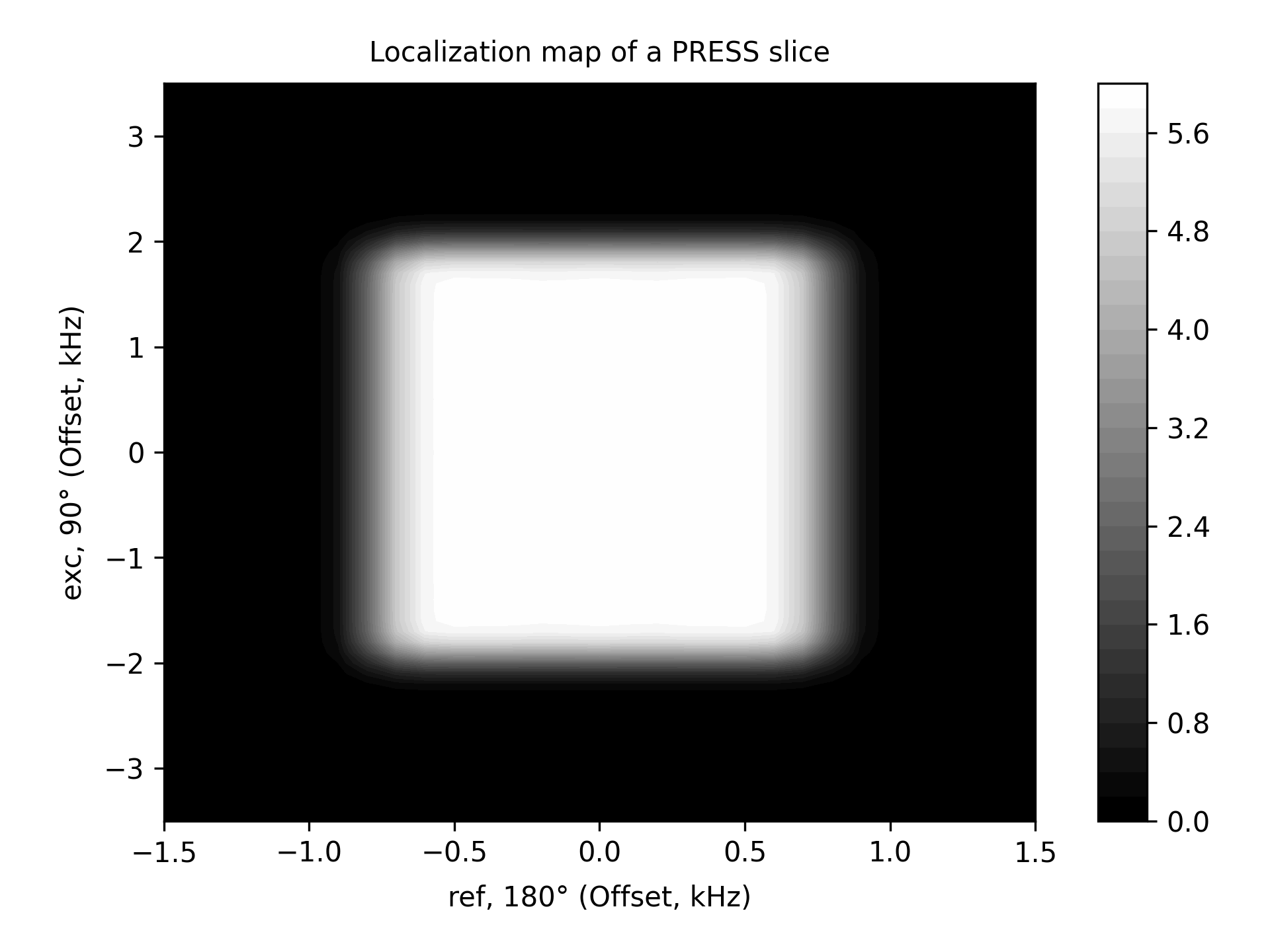
See Notes in the PRESS example for more explanations.
press2D (main input file)
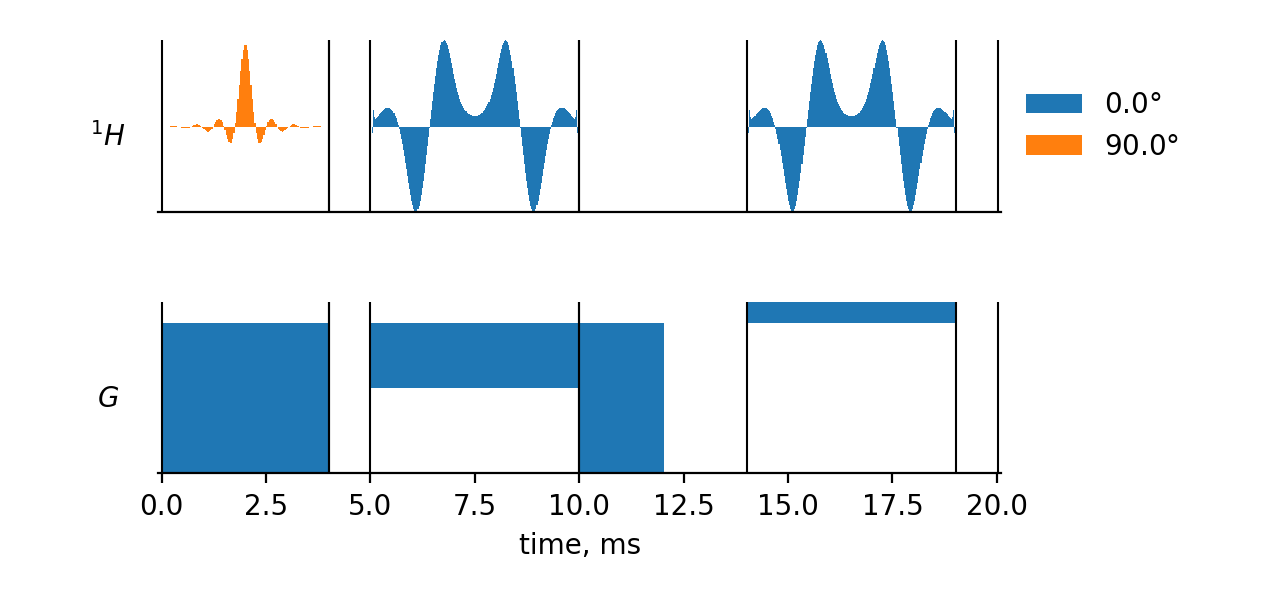
******************************************************************************* ** PRESS slice 2D map (based on the 'press' input file) ** Reference: Kaiser, L. G., Young, K., Matson, G. B. (2008) ** Numerical simulations of localized high field 1H MR spectroscopy. ** Journal of Magnetic Resonance, 195(1), 67–75. ****** The System ************************************************************* spectrometer(MHz) 125 spinning_freq(kHz) * channels H1 nuclei * atomic_coords * cs_isotropic MOL.cs ppm csa_parameters * j_coupling MOL.j quadrupole * dip_switchboard * csa_switchboard * exchange_nuclei * bond_len_nuclei * bond_ang_nuclei * tors_ang_nuclei * groups_nuclei * ******* Pulse Sequence ****************************************************** CHN 1 timing(usec) (t128.tm) 0 (0) (t200.tm) 0 (0 0) (t200.tm) 0 power(kHz) slr90.pwr 0 0 s180.pwr 0 0 0 s180.pwr 0 phase(deg) 90 0 0 0 0 0 0 0 0 freq_offs(kHz) 0 0 0 0 0 0 0 0 0 CHN G timing(usec) (t128.tm) 0 (0) (t200.tm) 0 (0 0) (t200.tm) 0 grad_offs(kHz) 0 0 0 0 0 0 0 0 0 ******* Variables *********************************************************** ** slr90.pwr is the selective excitation pulse powers, kHz, for a 10 ms pulse ** s180.pwr is the selective refocusing pulse powers, kHz, for a 10 ms pulse ** See examples/selective-pulses to investigate how these pulses work ** Specify selective pulses lengths, usec T90 = 4000 T180 = 5000 ** Specify (refocusing time)/(pulse length) ratio for the excitation pulse ** The refocusing time for the excitation pulse is given as a comment in slr90.pwr r90 = 0.5050 // slr90 ** Set up the selective excitation pulse pulse_[1 2]_1=T90/128 psf_1_1=10000/T90 ** Set up the selective refocusing pulses pulse_[1 2]_4=T180/200 pulse_[1 2]_7=T180/200 psf_1_[4 7]=10000/T180 ** Coherence pathway selected with the crusher gradinents select_[2 5]=[-1 1] ** Localization: slice 2D map ** ** G1, G2, G3 are offsets, kHz, generated by PFGs along X, Y, and Z G3 = 0.5 scan_par1d G2/-1.5:0.1:1.5/ scan_par2d G1/-3.5:0.1:3.5/ grad_offs_[1 4 7]=G[1 2 3] ** Refocus the linear phase generated by the selective excitation pulse pulse_[1 2]_6_1=T90*r90 grad_offs_6_1=G1 ** Additional interpulse delays (due to crusher gradients etc.) t1=1000 t2=1000 pulse_[1 2]_3_1=t1 pulse_[1 2]_6_2=t1+t2 pulse_[1 2]_8_1=t2 ** Take absolute value of the signal pdata_re=abs(complex(pdata_re,pdata_im)) x_label="ref, 180° (Offset, kHz)" y_label="exc, 90° (Offset, kHz)" fig_options="--contn 40 --cmap gray" fig_title="Localization map of a PRESS slice" ******* Options *************************************************************** rho0 F1z observables F1p EulerAngles * n_gamma * line_broaden(Hz) * zerofill * FFT_dimensions * options -re -macroMOL=myo-inositol -sysh6 -varppm_center_1=3.6 -py -contf ******************************************************************************* -- A different molecule can be easily chosen by using different values for the command line options -macroMOL=... -sysh... -varppm_center_1=...
Files referenced from the main input file:
mio-inositol.cs
3.5217 4.0538 3.5217 3.6144 3.2690 3.6144 0.1
mio-inositol.j
1 2 2.889 1 6 9.998 2 3 3.006 3 4 9.997 4 5 9.485 5 6 9.482
slr90.pwr
0.0000000e+00 -2.9240431e-05 -5.5617998e-05 -8.9752483e-05 -1.3153110e-04 ... -1.3153110e-04 -8.9752483e-05 -5.5617998e-05 -2.9240431e-05 0.0000000e+00
s180.pwr
0.0000000e+00 -1.6446000e-03 5.1245000e-03 2.8855000e-03 2.5472000e-03 ... 2.5488000e-03 2.8869000e-03 5.1258000e-03 -1.6435000e-03 0.0000000e+00
t128.tm
78.125 78.125 78.125 ... 78.125 78.125 78.125
t200.tm
50 50 50 ... 50 50 50
